State-Space Model#
We may wish to consider another Bayesian setting, but under a markovian setting that evolves over time, namely the latent state space model:
Where \(y\) is observed data, \(x\) is latent, and we wish to infer on \(\theta\)
We first compute the likelihood of y, given by
Now we construct the posterior
Using established result, we know
Where the posterior covariance is given by
Or equivalently in a more elegant form
And the posterior mean \(M\) is given by
or more elegantly
As an example, consider the following parameters:
And we observed \( y = \begin{pmatrix}1\\[3pt]1\end{pmatrix}. \)
In this case we know the posterior is $\( p(\theta\mid y) = \mathcal{N}(y; M, \Sigma) \)\( Where \)M, \Sigma\( depends on \)\epsilon$ and is computed as follow:
import numpy as np
import pandas as pd
m = np.array([[2], [0]])
P = np.eye(2)
C = np.array([[1, 1], [1, 1.01]])
Q = 0.01 * np.eye(2)
y = np.array([[1], [1]])
A_base = np.array([[1, 0], [0, 0]], dtype=float) # Base part of A
R_base = np.array([[1, 0], [0, 1]], dtype=float) # Base part of R
epsilons = np.logspace(-3, 0, num=4) # 10^-3, 10^-2, 10^-1, 10^0
post_var = []
post_means = []
for epsilon in epsilons:
A = A_base.copy()
A[1, 0] = epsilon # Update A with the varying epsilon
R = epsilon * R_base # Update R with the varying epsilon
S = A @ Q @ A.T + R
Sigma = np.linalg.inv(P + C.T @ A.T @ np.linalg.inv(S) @ A @ C)
post_var.append(np.round(Sigma, 4))
post_mean = Sigma @ (np.linalg.inv(P) @ m + C.T @ A.T @ np.linalg.inv(S) @ y)
post_means.append(np.round(post_mean.flatten(), 4)) # store 4 digits
# Create a DataFrame for visualization
df_post = pd.DataFrame(
{"Epsilon": epsilons,
"Posterior Mean": post_means,
"Posterior Variance": post_var}
)
df_post
| Epsilon | Posterior Mean | Posterior Variance | |
|---|---|---|---|
| 0 | 0.001 | [1.5032, -0.4968] | [[0.5027, -0.4973], [-0.4973, 0.5027]] |
| 1 | 0.010 | [1.5099, -0.4901] | [[0.505, -0.495], [-0.495, 0.505]] |
| 2 | 0.100 | [1.5681, -0.4319] | [[0.5258, -0.4742], [-0.4742, 0.5258]] |
| 3 | 1.000 | [1.6016, -0.3984] | [[0.6016, -0.3984], [-0.3984, 0.6016]] |
With \(\epsilon=1\), we have $$ p(\theta\mid y) = \mathcal{N}(y;
) $$
And we plot the posterior:
from scipy.stats import multivariate_normal
import matplotlib.pyplot as plt
# Define grid for contour plot
x_bb = np.linspace(-2, 5, 100)
y_bb = np.linspace(-4, 3, 100)
X_bb, Y_bb = np.meshgrid(x_bb, y_bb)
fig, axes = plt.subplots(1, 4, figsize=(20, 5))
plt.rcParams.update({'font.size': 12}) # Set font size
for i in df_post.index:
eps = df_post['Epsilon'][i]
post_mean = df_post['Posterior Mean'][i]
post_var = df_post['Posterior Variance'][i]
rv = multivariate_normal(mean=post_mean, cov=post_var)
# Evaluate the posterior density on the grid
pos = np.dstack((X_bb, Y_bb))
Z_bb = rv.pdf(pos)
# Plot the contour
ax = axes[i]
cnt = ax.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
ax.set_title(f'$\\epsilon = {eps:.3g}$')
ax.set_xlabel('$\\theta_1$')
ax.set_ylabel('$\\theta_2$')
fig.colorbar(cnt, ax=axes, orientation='vertical', fraction=0.02, pad=0.04, label='Density')
plt.suptitle('Target Distribution for Posterior $p(\\theta | y)$ for Different $\\epsilon$ Values')
plt.show()
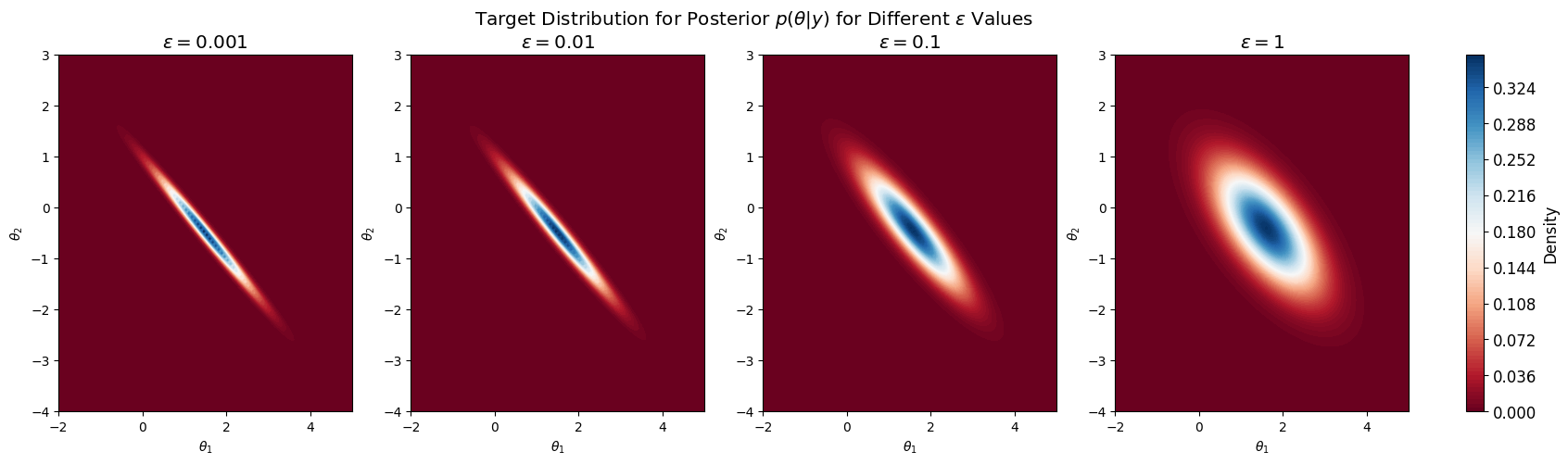
Now consider \(\epsilon=1\).
We can sample from the posterior if we know the analytical form.
Assume we don’t
