HMM with Example#
Consider the latent state space model:
Where \(y\) is observed data, \(x\) is latent, and we wish to infer on \(\theta\)
We first compute the likelihood of y, given by
Now we construct the posterior
Using established result, we know
Where the posterior covariance is given by
Or equivalently in a more elegant form
And the posterior mean \(M\) is given by
or more elegantly
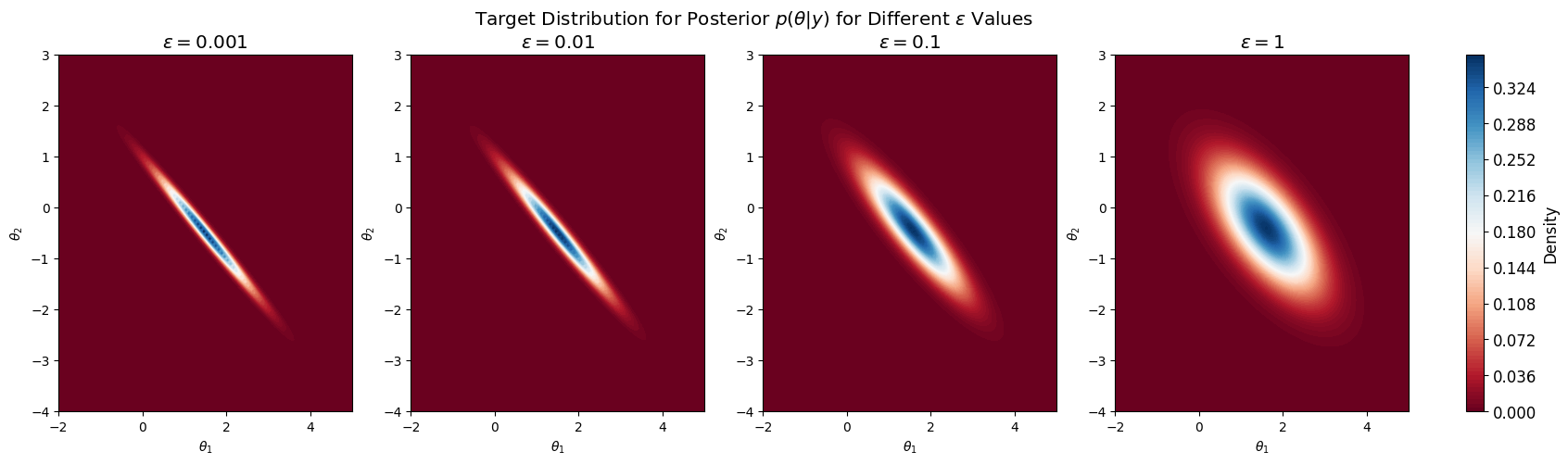
As an example, consider the following parameters:
And we observed \( y = \begin{pmatrix}1\\[3pt]1\end{pmatrix}. \)
In this case we know the posterior is
Where \(M, \Sigma\) depends on \(\epsilon\) and is computed as follow:
import numpy as np
import pandas as pd
m = np.array([[2], [0]])
P = np.eye(2)
C = np.array([[1, 1], [1, 1.01]])
Q = 0.01 * np.eye(2)
y = np.array([[1], [1]])
A_base = np.array([[1, 0], [0, 0]], dtype=float) # Base part of A
R_base = np.array([[1, 0], [0, 1]], dtype=float) # Base part of R
epsilons = np.logspace(-3, 0, num=4) # 10^-3, 10^-2, 10^-1, 10^0
post_var = []
post_means = []
for epsilon in epsilons:
A = A_base.copy()
A[1, 0] = epsilon # Update A with the varying epsilon
R = epsilon * R_base # Update R with the varying epsilon
S = A @ Q @ A.T + R
Sigma = np.linalg.inv(P + C.T @ A.T @ np.linalg.inv(S) @ A @ C)
post_var.append(np.round(Sigma, 4))
post_mean = Sigma @ (np.linalg.inv(P) @ m + C.T @ A.T @ np.linalg.inv(S) @ y)
post_means.append(np.round(post_mean.flatten(), 4)) # store 4 digits
# Create a DataFrame for visualization
df_post = pd.DataFrame(
{"Epsilon": epsilons,
"Posterior Mean": post_means,
"Posterior Variance": post_var}
)
df_post
| Epsilon | Posterior Mean | Posterior Variance | |
|---|---|---|---|
| 0 | 0.001 | [1.5032, -0.4968] | [[0.5027, -0.4973], [-0.4973, 0.5027]] |
| 1 | 0.010 | [1.5099, -0.4901] | [[0.505, -0.495], [-0.495, 0.505]] |
| 2 | 0.100 | [1.5681, -0.4319] | [[0.5258, -0.4742], [-0.4742, 0.5258]] |
| 3 | 1.000 | [1.6016, -0.3984] | [[0.6016, -0.3984], [-0.3984, 0.6016]] |
With \(\epsilon=1\), we have $$ p(\theta\mid y) = \mathcal{N}(y;
) $$
And we plot the posterior:
from scipy.stats import multivariate_normal
import matplotlib.pyplot as plt
# Define grid for contour plot
x_bb = np.linspace(-2, 5, 100)
y_bb = np.linspace(-4, 3, 100)
X_bb, Y_bb = np.meshgrid(x_bb, y_bb)
fig, axes = plt.subplots(1, 4, figsize=(20, 5))
plt.rcParams.update({'font.size': 12}) # Set font size
for i in df_post.index:
eps = df_post['Epsilon'][i]
post_mean = df_post['Posterior Mean'][i]
post_var = df_post['Posterior Variance'][i]
rv = multivariate_normal(mean=post_mean, cov=post_var)
# Evaluate the posterior density on the grid
pos = np.dstack((X_bb, Y_bb))
Z_bb = rv.pdf(pos)
# Plot the contour
ax = axes[i]
cnt = ax.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
ax.set_title(f'$\\epsilon = {eps:.3g}$')
ax.set_xlabel('$\\theta_1$')
ax.set_ylabel('$\\theta_2$')
fig.colorbar(cnt, ax=axes, orientation='vertical', fraction=0.02, pad=0.04, label='Density')
plt.suptitle('Target Distribution for Posterior $p(\\theta | y)$ for Different $\\epsilon$ Values')
plt.show()
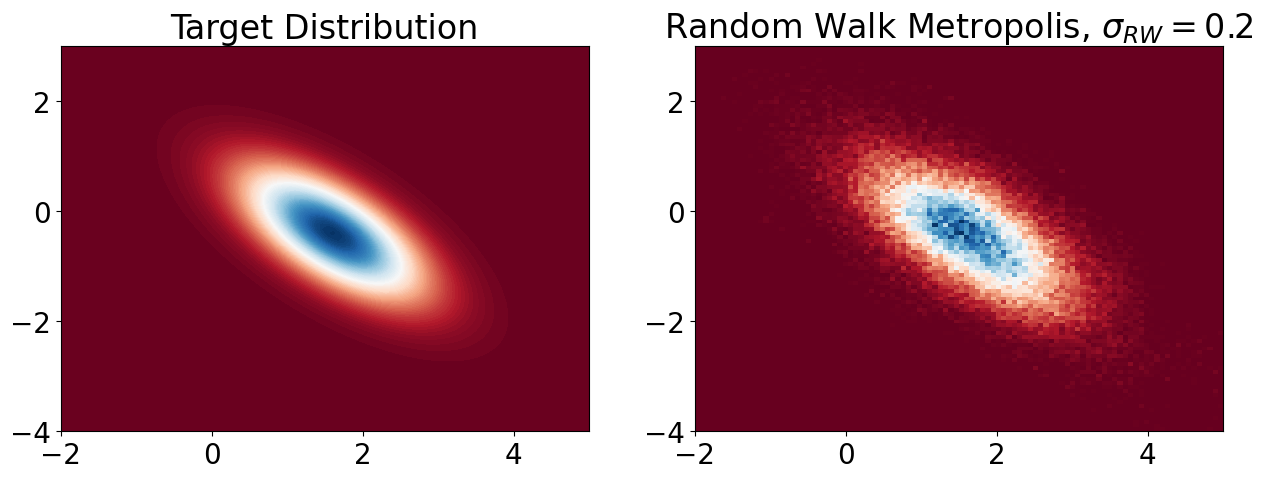
Random Walk Metropolis Hastings (RWMH)#
Now consider \(\epsilon=1\). Recall that we have access to the true posterior:
Suppose we only have access to the unnormalised posterior:
We can use RWMH to sample from the posterior \(p(\theta | y)\)
I use \(x\) to denote \(\theta\), and use \(x_s\) to denote a new state for simplicity. My RWMH algorithm follows these steps:
Initialize at \(x_0 = (0, 0)\), fix \(\sigma_{\text{RW}}\)
For each iteration:
Propose a new state under \(q(\theta' | \theta_{n-1}) = \mathcal{N}(\theta_{n-1}, \sigma_{\text{rw}}) \) $\( x_s = x_{n-1} + \sigma_{\text{rw}} \cdot \mathcal{N}(0, I) \)$
Compute acceptance probability:
$\( a = \min\left(1, \frac{\pi(x_s)}{\pi(x_{n-1})} \right) \)$Accept or reject:
With probability \(a\), set \(x_n = x_s \).
Otherwise, set \( x_n = x_{n-1} \).
I set \(\sigma_{\text{rw}} = 0.2 \).
If it’s too small, proposals are close to the current state → slow exploration.
Otherwise if too large, proposals often land in low-probability regions → high rejection rate.
# Set random seed
rng = np.random.default_rng(24)
# Load posterior mean and variance
post_mean = df_post['Posterior Mean'][3]
post_var = df_post['Posterior Variance'][3]
rv = multivariate_normal(mean=post_mean, cov=post_var)
# Define log of the new target distribution
def logPi(x):
return rv.logpdf(x)
# MCMC Sampling
def RWMH(logPi, sigma_rw = 0.5, N = 100000, burnin = 1000):
samples_RW = np.zeros((2, N))
samples_RW[:, 0] = np.array([0, 0])
# Random Walk Metropolis-Hastings Algorithm
for n in range(1, N):
# Proposal sample from Gaussian random walk
x_s = samples_RW[:, n-1] + sigma_rw * rng.normal(size=2)
# Metropolis-Hastings acceptance step
u = rng.uniform(0, 1)
if np.log(u) < logPi(x_s) - logPi(samples_RW[:, n-1]):
samples_RW[:, n] = x_s
else:
samples_RW[:, n] = samples_RW[:, n-1]
return samples_RW
burnin = 1000
samples_RW = RWMH(logPi, sigma_rw = 0.2, burnin=burnin)
# Visualization
x_bb = np.linspace(-2, 5, 100)
y_bb = np.linspace(-4, 3, 100)
X_bb, Y_bb = np.meshgrid(x_bb, y_bb)
pos = np.dstack((X_bb, Y_bb))
Z_bb = rv.pdf(pos)
plt.figure(figsize=(15, 5))
plt.rcParams.update({'font.size': 20})
# Contour plot of target distribution
plt.subplot(1, 2, 1)
plt.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
plt.title('Target Distribution')
# Histogram of sampled points
plt.subplot(1, 2, 2)
plt.hist2d(samples_RW[0, burnin:], samples_RW[1, burnin:],
100, cmap='RdBu', range=[[-2, 5], [-4, 3]], density=True)
plt.title('Random Walk Metropolis, $\\sigma_{RW} = 0.2$')
plt.show()
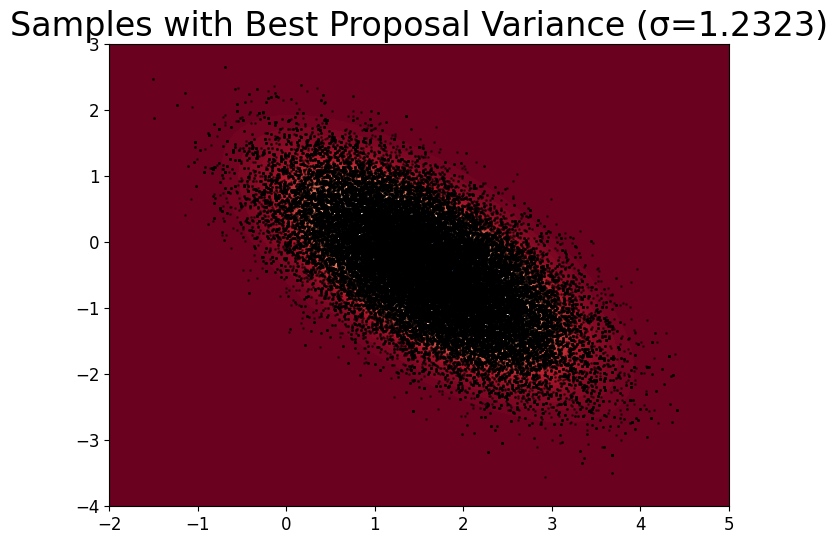
Finding best proposal variance#
We tune proposal variance \(\sigma^2_{MH}\) between \(10^{-4}\) to 4, and run RWMH with each variance.
import statsmodels.api as sm
sigma_values = np.logspace(-4, np.log10(4), num=10)
acf_results = {} # Store results
samples_results = {}
for sigma_rw in sigma_values:
samples_RW = RWMH(logPi, sigma_rw=sigma_rw, burnin=burnin)
# Store results
acf_results[sigma_rw] = sm.tsa.acf(samples_RW[0, burnin:], nlags=50)
samples_results[sigma_rw] = samples_RW
# Find the best sigma_rw (lowest autocorrelation at lag 1)
best_sigma_rw = min(acf_results, key=lambda s: acf_results[s][1])
samples_RW_best = samples_results[best_sigma_rw]
print(f'Best sigma_rw: {best_sigma_rw}')
# Visualization
x_bb = np.linspace(-2, 5, 100)
y_bb = np.linspace(-4, 3, 100)
X_bb, Y_bb = np.meshgrid(x_bb, y_bb)
pos = np.dstack((X_bb, Y_bb))
Z_bb = rv.pdf(pos)
plt.figure(figsize=(8, 6))
plt.tick_params(axis='y', labelsize=12)
plt.tick_params(axis='x', labelsize=12)
plt.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
plt.scatter(samples_RW_best[0, burnin:], samples_RW_best[1, burnin:],
s=1, color='black', alpha=0.5)
plt.title(f'Samples with Best Proposal Variance (σ={best_sigma_rw:.4f})')
plt.xlim(-2, 5)
plt.ylim(-4, 3)
plt.show()
Best sigma_rw: 1.2323100555166684
Importance Sampler for Posterior#
Suppose we don’t even have access to the likelihood \(p(y | \theta)\).
We know that
Where \( q(x \mid \theta) \) is our proposal
Hence we can sample \(\{X_i\}_{i=1}^N\) from \( q(x \mid \theta) \), then compute $\( \frac{1}{N} \sum_{i=1}^N p(y \mid X_i)\; \frac{p(X_i \mid \theta)}{q(X_i \mid \theta)} \)\( as an unbiased estimator of \)p(y \mid \theta)$
We can also denote our estimator as $\( \sum_{i=1}^N w_i \; p(y \mid X_i) \)$
Where \( w_i = \frac{1}{N} \frac{p(X_i \mid \theta)}{q(X_i \mid \theta)} \) is the self-normalizing weight
# We re-define parameters again
epsilon = 1
theta = [0, 0]
y = [1, 1]
m = np.array([2, 0])
Q = np.array([[0.01, 0], [0, 0.01]])
P = np.array([[1, 0], [0, 1]])
C = np.array([[1, 1], [1, 1.01]])
A = np.array([[1, 0], [epsilon, 0]])
R = np.array([[epsilon, 0], [0, epsilon]])
def log_p_y_given_theta(y, theta):
# true log density of y given theta
mean = np.dot(A, np.dot(C, theta))
cov = np.dot(A, np.dot(Q, A.T)) + R
diff = y - mean
return (-0.5 * np.dot(diff.T, np.linalg.inv(cov)).dot(diff)
- 0.5 * np.log(np.linalg.det(2 * np.pi * cov)))
def log_p_x_given_theta(x, theta):
diff = x - np.dot(C, theta)
return (-0.5 * np.dot(diff.T, np.linalg.inv(Q)).dot(diff)
- 0.5 * np.log(np.linalg.det(2 * np.pi * Q)))
def log_p_y_given_x(y, x):
diff = y - np.dot(A, x)
return (-0.5 * np.dot(diff.T, np.linalg.inv(R)).dot(diff)
- 0.5 * np.log(np.linalg.det(2 * np.pi * R)))
def log_p_theta(theta):
diff = theta - m
return (-0.5 * np.dot(diff.T, np.linalg.inv(P)).dot(diff)
- 0.5 * np.log(np.linalg.det(2 * np.pi * P)))
def logq(x, sig): # log-density of the proposal
# proposal N(0, sig^2) in 2D
return (- x[0]**2/(2 * sig**2) - x[1]**2/(2 * sig**2)
- np.log(2 * np.pi * sig**2))
# Self-normalised importance sampling of p(y|theta)
def SNIS_for_loop(N, sig, theta=[0, 0]):
'''Classical SNIS using a for loop'''
# we used that y is fixed
x_samples = rng.normal(0, sig, (N, 2)) # propose x
logw = np.zeros(N) # log-weights
phi_X = np.zeros(N)
for i in range(N):
logw[i] = (log_p_x_given_theta(x_samples[i, :], theta)
- logq(x_samples[i, :], sig))
phi_X[i] = np.exp(log_p_y_given_x(y, x_samples[i, :]))
logw = logw - np.max(logw) # to avoid numerical issues
w = np.exp(logw)/np.sum(np.exp(logw)) # normalising the weights
return np.dot(w, phi_X)
We also give a faster IS algorithm below, where we use vectorization to avoid the time-consuming for loop
In particular, since we know that \( \epsilon = 1\) and \( y = (1, 1) \) are fixed, we have the likelihood
We implement this in a vectorized way below
def p_y_given_x(x):
"""Compute the density of N((1,1); (x1, x1), I_2) for given x (N,2)."""
mu = np.array([1, 1]) # Mean
norm_const = 1 / (2 * np.pi) # (1 / sqrt((2π)^2 * det(I_2)))
diff = x - mu # (N,2)
exp_term = np.exp(-0.5 * np.sum(diff**2, axis=1)) # (N,)
return norm_const * exp_term # (N,)
def q(x, sig):
# x has shape (N, 2)
return multivariate_normal.pdf(x, mean=np.zeros(2), cov=sig**2 * np.eye(2))
def p_x_given_theta(x, theta):
# x has shape (N, 2)
return multivariate_normal.pdf(x, mean=np.dot(C, theta), cov=Q)
# we provide a new definition of IS with vectorization
def SNIS(N, sig, theta=[0, 0]):
x_samples = rng.normal(0, sig, (N, 2)) # propose x
# y | x likelihood
phi_X = p_y_given_x(x_samples)
# element wise division, w is (N, )
w = p_x_given_theta(x_samples, theta) / q(x_samples, sig)
return np.mean(w * phi_X)
# check result with SNIS with example run with sig=0.1
K, sig_ = 10000, 0.1
phi_SNIS = SNIS_for_loop(K, sig_ )
print('SNIS for loop:', phi_SNIS)
phi_SNIS_vec = SNIS(K, sig_)
print('SNIS vectorized:', phi_SNIS_vec)
true_lik = np.exp(log_p_y_given_theta(y, theta)) # true p(y|theta)
print('True likelihood:', true_lik)
SNIS for loop: 0.0590472992284872
SNIS vectorized: 0.058644608053991326
True likelihood: 0.05912091432281943
# Relative Absolute Error
def RAE(est):
return np.abs(est - true_lik)/true_lik
Pseudo-marginal Metropolis Hastings (PMMH)#
We can plug this IS estimator into the MH ratio to obtain pseudo-marginal Metropolis Hastings (PMMH)
In our case we have
where we assume
is intractable, and can be replaced by the IS estimator \( \hat{p}(y|\theta) \) as described above (in 1.6 & 1.7)
We know \( p(y , x \mid \theta) \) can be computed pointwise for any \( x, \theta \), since \( y \) is the fixed data.
Hence we can plug the IS estimator \( \hat{p}(y|\theta) \) into the MH ratio to obtain the PMMH:
Initialize at \(\theta_0 = (0, 0)\), fix \(\sigma_{\text{MH}}\) and \(\sigma_{\text{IS}}\)
For each iteration:
Propose a new state under \(q(\theta' | \theta_{n-1}) = \mathcal{N}(\theta_{n-1}, \sigma_{\text{rw}}) \)
\[ \theta' = \theta_{n-1} + \sigma_{\text{MH}} \cdot \mathcal{N}(0, I) \]Run IS to compute likelihood under \(\theta'\): \(\hat{p}(y|\theta')\) using the algo described above
Compute acceptance probability:
$\( \alpha(\theta_{n-1}, \theta') = \min \left( 1, \frac{\hat{p}(y|\theta') p(\theta')(\theta_{n-1} | \theta')}{\hat{p}(y|\theta_{n-1}) p(\theta_{n-1}) q(\theta' | \theta{n-1})} \right) \)$
Note if we are using symmetric proposal \( q(\theta' | \theta_{n-1}) = q(\theta_{n-1} | \theta')\), then the ratio becomes $\( \alpha(\theta_{n-1}, \theta') = \min \left( 1, \frac{\hat{p}(y|\theta') p(\theta')}{\hat{p}(y|\theta_{n-1}) p(\theta_{n-1}) } \right) \)$
Accept or reject:
With probability \(a\), set \(\theta_n = \theta' \).
Otherwise, set \( \theta_n = \theta_{n-1} \).
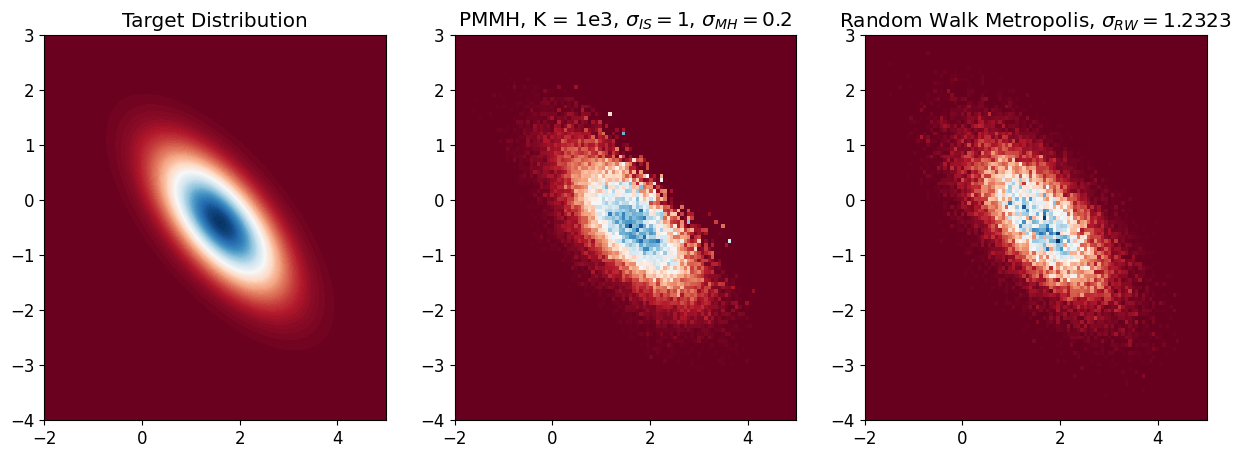
After tuning, we find out that sig_IS=1, sig_MH=0.2 leads to the best result
import statsmodels.api as sm
def PMMH(logPi, sig_IS, sig_MH, K = 10,
N = 100000, burnin = 5000, use_for_loop=False):
# logPi is the prior logp(theta)
sample_MH = np.zeros((2, N))
sample_MH[:, 0] = np.array([0, 0])
if use_for_loop:
IS_lik = np.log(SNIS_for_loop(K, sig_IS, theta=sample_MH[:, 0]))
else:
IS_lik = np.log(SNIS(K, sig_IS, theta=sample_MH[:, 0]))
for n in range(1, N):
# Proposal sample from Gaussian random walk
x_s = sample_MH[:, n-1] + sig_MH * rng.normal(size=2)
# IS estimate of the log likelihood
IS_lik_new = log_p_y_given_theta(y, x_s)
# print('true', IS_lik_new)
IS_lik_new = np.log(SNIS(K, sig_IS, theta=x_s))
# print(IS_lik_new)
ratio = (IS_lik_new - IS_lik + logPi(x_s) - logPi(sample_MH[:, n-1]))
# Metropolis-Hastings acceptance step
u = rng.uniform(0, 1)
if np.log(u) < ratio:
sample_MH[:, n] = x_s
IS_lik = IS_lik_new
else:
sample_MH[:, n] = sample_MH[:, n-1]
return sample_MH[:, burnin:]
acf_K = {} # Store acf
def plot_MH(k, sig_IS, sig_MH, use_for_loop=False):
samples_MH = PMMH(log_p_theta, sig_IS=sig_IS, sig_MH=sig_MH,
K = k, N = 100000, burnin=5000, use_for_loop=use_for_loop)
acf_K[k] = sm.tsa.acf(samples_MH[0, :], nlags=50)
plt.figure(figsize=(15, 5))
plt.rcParams.update({'font.size': 20})
# Contour plot of target distribution
plt.subplot(1, 2, 1)
plt.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
plt.title('Target Distribution')
# Histogram of sampled points
plt.subplot(1, 2, 2)
plt.hist2d(samples_MH[0, :], samples_MH[1, :],
100, cmap='RdBu', range=[[-2, 5], [-4, 3]], density=True)
plt.title(f'Pseudo Metropolis, K = {k}')
plt.show()
return samples_MH
samples_MH = plot_MH(1000, 1, 0.2, use_for_loop=True)
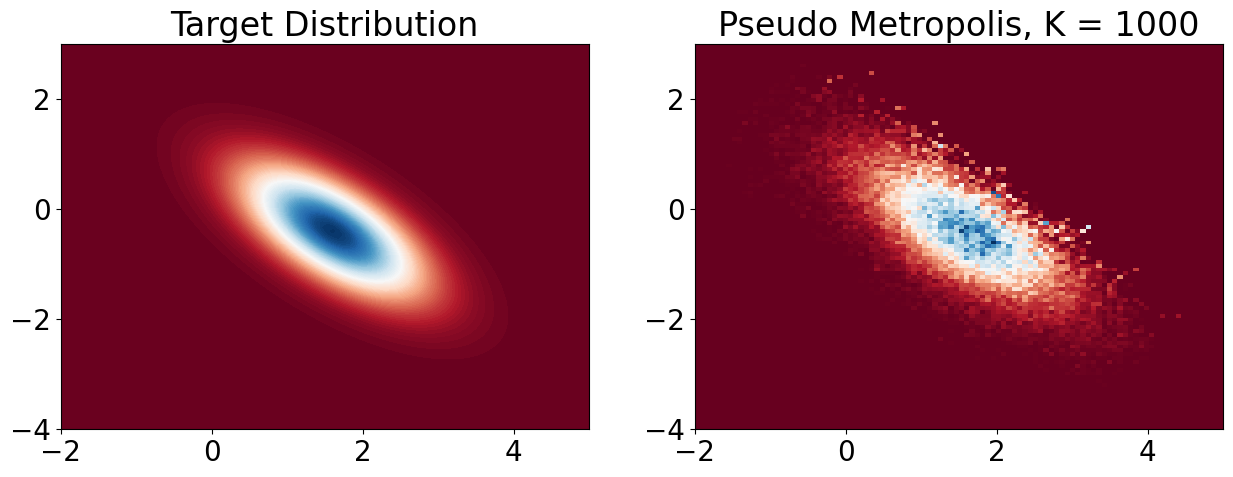
samples_MH = plot_MH(1000, 1, 0.2, use_for_loop=True)
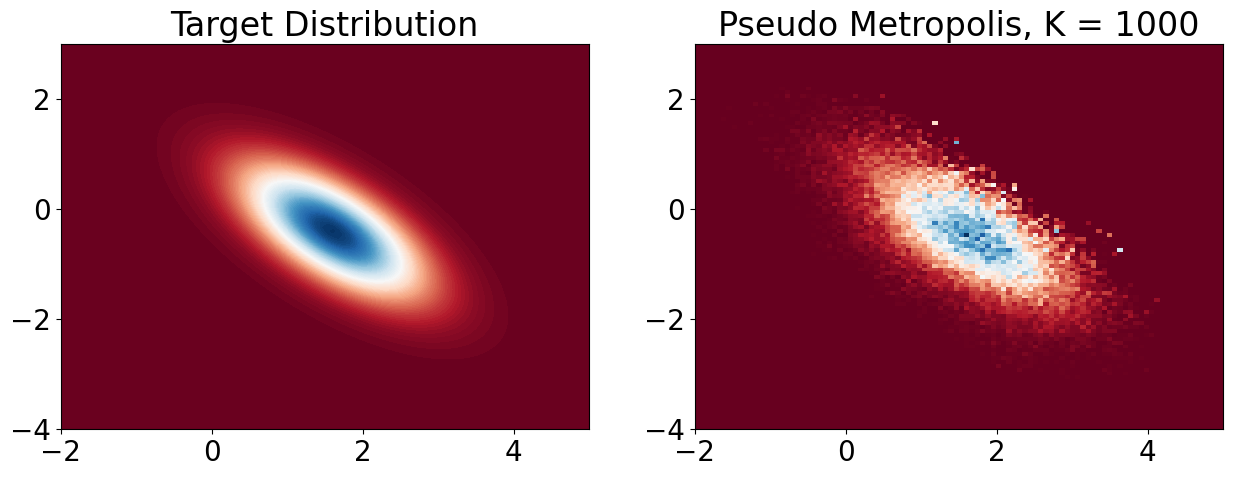
# compare K = 1000 with RWMH
plt.rcParams.update({'font.size': 12}) # Set font size
plt.figure(figsize=(15, 5))
# Contour plot of target distribution
plt.subplot(1, 3, 1)
plt.contourf(X_bb, Y_bb, Z_bb, 100, cmap='RdBu')
plt.title('Target Distribution')
# Histogram of sampled points
plt.subplot(1, 3, 2)
plt.hist2d(samples_MH[0, :], samples_MH[1, :],
100, cmap='RdBu', range=[[-2, 5], [-4, 3]], density=True)
plt.title('PMMH, K = 1e3, $\\sigma_{IS} = 1$, $\\sigma_{MH} = 0.2$')
plt.subplot(1, 3, 3)
plt.hist2d(samples_RW_best[0, burnin:], samples_RW_best[1, burnin:],
100, cmap='RdBu', range=[[-2, 5], [-4, 3]], density=True)
plt.title('Random Walk Metropolis, $\\sigma_{RW} = 1.2323$')
plt.show()